Early tool could predict life-threatening complications
Despite its crucial role in pregnancy, the placenta is a highly understudied organ. Evaluation of the placenta after birth can provide timely information about the health of both mother and baby, yet the vast majority of placentas are discarded without analysis, as pathological examinations can be costly, time-intensive, and require specialized expertise.
But what if the placenta could be analyzed with simple photographs? Researchers at The Pennsylvania State University (Penn State) are developing a tool that uses AI to evaluate a picture of the placenta, providing a prediction for multiple adverse outcomes, such as infection or sepsis. With further refinement, this technology could flag individual patients for enhanced monitoring or immediate follow-up care, potentially improving infant and maternal outcomes, especially in low-income regions. Their research was recently featured on the cover of Patterns, a journal published by Cell.
“Infant mortality is the highest in the first hours and days after birth, and while placental pathology exams can provide a wealth of diagnostic information, only about 20% of placentas are analyzed in the United States,” said study author Alison Gernand, Ph.D., an associate professor of nutritional sciences at Penn State. “We want to make it easier to study the placenta, and ultimately, we want to develop a tool that can analyze the placenta to impact clinical care.”
Pairing pathology reports with placental photographs
As a first step in the development of their tool, the researchers needed to correlate photographs of the placenta with their respective pathology reports to understand which features might be prognostic. They trained their tool by analyzing paired photographs and pathology reports, using both an image processing algorithm and a local large language model (LLM) to tease out patterns in the data. This approach is called vision-and-language contrastive learning.
Traditional AI image analysis revolves around one prediction—a single diagnosis or outcome that is extrapolated from the information embedded in the image. “The advantage of using a contrastive learning approach is that you can take the entire pathology report and the corresponding medical photographs, encode them into the model, and then the AI can learn the associations between the text and the images,” explained study author James Wang, Ph.D., a distinguished professor of information sciences and technology at Penn State. “In this way, we are able to correlate placental features that correspond with multiple different diagnoses described in the pathology reports.”
To develop their model, the researchers used approximately 30,000 image-report pairs collected at Northwestern Memorial Hospital in Chicago. This initial step “taught” the image processing algorithm about specific medical information contained in the pathology reports. Next, the researchers fed the model roughly 1,500 image-report pairs (collected from the same hospital but not used previously) so that it could extract image features and predict specific clinical outcomes. The model was then validated using another set of image-report pairs (approximately 1,500).
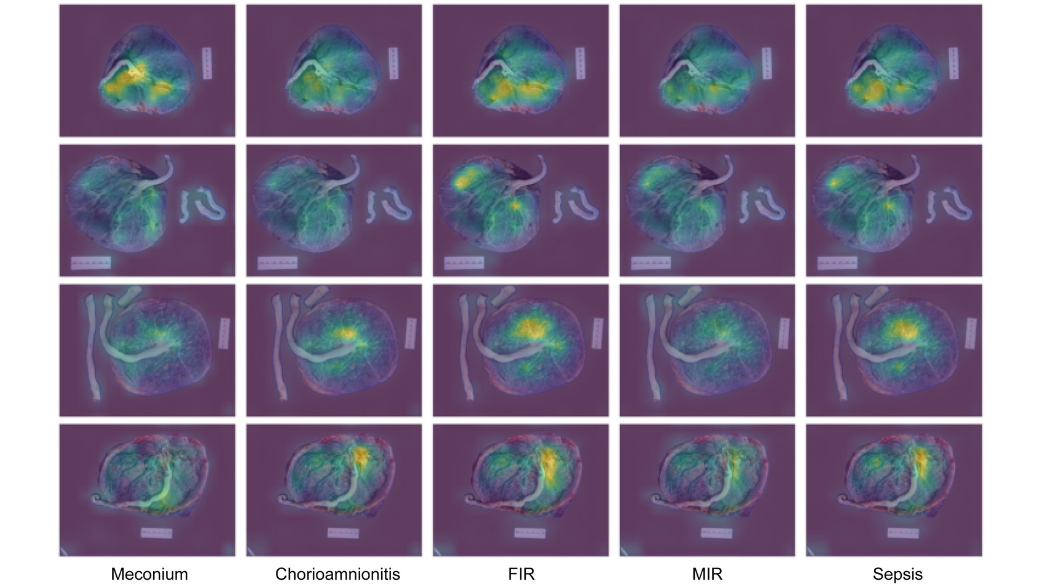
The researchers found that their model could identify a host of conditions, including sepsis (94% of the time), the presence of meconium (87%), fetal and maternal inflammatory responses (86% and 78%, respectively), and chorioamnionitis (76%).
“While these conditions are pathologically distinct, they are somewhat interrelated,” explained Gernand. “Down the road, we’d like to expand our model to include additional diagnoses, because there are a lot of different pathologies that can be identified in the placenta, and some are connected to immediate health risks such as postpartum hemorrhage, which can be life-threatening.”
Focusing on image quality
The images collected at Northwestern Memorial Hospital were captured with a dedicated pathology specimen photography system, which includes a fixed high-resolution camera and optimal lighting. However, not all hospitals have access to such technology.
To understand how their technique might perform in a different setting, the researchers tested their model using 353 image-report pairs previously collected from the Mbarara Regional Referral Hospital in Mbarara, Uganda. Photographs collected here were captured using a handheld and off-the-shelf digital camera.
While the model was less robust in this setting, it could still predict the presence of meconium and fetal and maternal inflammatory responses. Neither chorioamnionitis nor sepsis were included in the pathology reports, so these conditions could not be predicted within this context.
“Because the image quality decreased between the two locations, we weren’t surprised that the performance of our model also decreased,” explained first study author Yimu Pan, an informatics doctoral candidate in the Wang lab. “We expect that we can improve the model’s accuracy by incorporating training images from multiple sites using different types of cameras, which will help ensure that the model can perform well across different locations.”
The research team also evaluated how specific image artifacts could affect the performance of their model, analyzing factors such as blood spots, shadows, or color intensity. To do this, they artificially distorted the images and intentionally introduced them into the model’s internal validation data. They found that several artifacts markedly reduced the performance of their model, including glare, using JPEG compression to reduce file sizes, and blur. In contrast, other artifacts, such as shadows, had a less significant impact. “These findings can supply guidance for clinical photography, providing insight into factors that can affect the performance of future imaging models,” noted Pan.
Zooming out and looking ahead
While the preliminary model has promising applications, the authors note that there are a few limitations. Notably, the model performed significantly better among mothers aged 45 and above. “The placentas collected at Northwestern Memorial Hospital were analyzed because there was an issue with the pregnancy, and older mothers are more likely to experience pregnancy complications,” explained Gernand. “In this way, our dataset had an artificially high age range compared to the general obstetric population.” Future work will include incorporating additional data from younger mothers, she said.
Further, the model in this study used only fetal-side placental images. The researchers hope to incorporate images of the placenta taken from the maternal side, which can provide additional diagnostic information.
“This resourceful technology could someday improve existing workflows and help clinicians triage their patients more efficiently,” said Rui Carlos Pereira de Sá, Ph.D., a program director in the Division of Health Informatics Technologies at NIBIB. “A tool like this one has the potential to improve maternal and infant outcomes everywhere, especially in regions without access to state-of-the-art diagnostic technologies.”
This study was funded by a grant from NIBIB (R01EB030130). The principal investigators on this grant are Gernand, Wang, and Jeffrey Goldstein, M.D., Ph.D., a perinatal pathologist at Northwestern Memorial Hospital. The external validation data came from a study supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development under awards R01HD112302 and K23AI138856.
Study reference: Yimu Pan, et al. Cross-modal contrastive learning for unified placenta analysis using photographs. Patterns, Volume 5, Issue 12, 2024. https://doi.org/10.1016/j.patter.2024.101097
About the graphics: These images originally appeared in the Patterns publication which is available under the the Creative Commons CC-BY-NC-ND license.
