A team of researchers has established a ribonucleic acid (RNA)-based method that drives cells in the body to produce therapeutic proteins and secrete them into the bloodstream. The approach could potentially extend the lifespan of drugs in the body, reducing the burden on patients who require frequent drug administrations.
The researchers, based at UT Southwestern Medical Center, leveraged a naturally occurring biomolecule called a signal peptide that determines where cells send proteins — acting like a shipping label — to release proteins into the blood that would normally remain in the cell. In a report published in Proceedings of the National Academy of Sciences, the new approach enabled secretion and increased the circulation time of a therapeutic protein compared to standard injections in a mouse model of psoriasis, demonstrating beneficial effects in these mice and separately, in animal models of cancer.
The modular nature of the technique suggests that it could be adapted to address a wide range of diseases.
“This could potentially be a powerful platform technology. By linking a signal peptide to any particular protein — insulin for patients with diabetes for example — you could very easily tailor this technique to different disease conditions,” said Jermont Chen, Ph.D., a program director in the National Institute of Biomedical Imaging and Bioengineering (NIBIB) Division of Discovery Science and Technology.
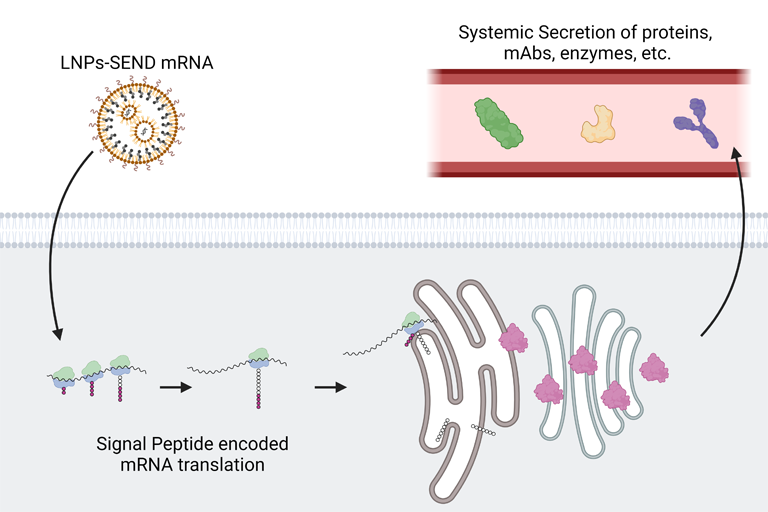
Messenger RNA, or mRNA, is a biomolecule that provides instructions for cellular machinery to produce proteins. Scientists have managed to introduce mRNA into the body to get cells to make particular proteins, such as with mRNA vaccines. But with this approach, proteins are usually trapped inside of the cell. With autoimmune disorders, some cancers, and many other diseases, therapeutic proteins perform best when they circulate throughout the body via the bloodstream.
Seeking to break therapeutic proteins out of their cellular confines, Daniel Siegwart, Ph.D., a professor of biomedical engineering and biochemistry, took inspiration from natural proteins that are normally found in the bloodstream.
Blood proteins produced by the liver feature small structures at their tail ends called signal peptides, which determine a protein’s destination. Some of these peptides instruct cells to shuttle proteins to the cellular membrane or nucleus, while others cause proteins to be delivered to a cellular organelle called the endoplasmic reticulum (ER), where they are prepared for release into circulation.
“We wanted to know if we could copy and paste specific signal peptides from secreted proteins to other proteins that normally remain in the cell, to get them into the bloodstream. If that were feasible, then we could create a factory in the body to produce and secrete medicine,” said Siegwart, corresponding author of the study.
The authors of the study designed mRNA sequences that would attach a signal peptide from a blood clotting protein onto the ends of other proteins. They packaged sequences into special nanoparticles the team previously designed to deliver mRNA to the liver.
In one set of experiments, the authors tested their approach, named Signal peptide Engineered Nucleic acid Design, or SEND, in mice with a psoriasis-like skin condition.
Psoriasis can be treated with an injection of a synthetic protein called etanercept. To evaluate their SEND platform, the researchers generated an mRNA construct that directed cells to make a version of etanercept that includes a signal peptide to navigate the secretion process through the ER. Once the therapeutic protein reaches the ER, the signal peptide is cleaved, allowing the cell to release an identical version of the etanercept medicine into the bloodstream.
For comparison, the researchers intravenously injected etanercept in one set of mice and SEND mRNA-filled nanoparticles in another group. They then recorded how long the protein stayed in the blood in each group.
They found that, when injected directly, the protein circulated for less than 24 hours, whereas proteins produced via SEND lasted nearly a week. Over the course of the experiment, the animals’ own cells produced roughly 12 times more etanercept than the amount injected into mice in the other group.
The skin of SEND-treated mice also recovered significantly compared to the skin of animals receiving nanoparticles carrying the signal peptide but no therapeutic protein.
In another set of experiments, the authors tested the effects of SEND in mouse models of two different types of cancer, adenocarcinoma and melanoma. This time, they linked the same signal peptide to the anticancer drug anti-PDL1.
Compared to treatment with nanoparticles that lacked mRNA to produce the drug, the full SEND treatment both reduced tumor growth and extended the animals’ survival for the two cancer types.
Now, the authors have their sights set on additional applications, such as enzyme replacement therapy, and larger animal models to bring the SEND platform closer to the clinic.
“We imagine that, if translated to humans, this platform could allow someone to receive anti-inflammatory medication once a month instead of a few times a week, for example,” said Siegwart. “And since the COVID-19 vaccine established a new model of health care deployment for mRNA-based medication, this all could potentially be done at the pharmacy or at home.”
This research was supported by grants from NIBIB (R01EB025192) and the National Cancer Institute (P30CA142543).
This science highlight describes a basic research finding. Basic research increases our understanding of human behavior and biology, which is foundational to advancing new and better ways to prevent, diagnose, and treat disease. Science is an unpredictable and incremental process—each research advance builds on past discoveries, often in unexpected ways. Most clinical advances would not be possible without the knowledge of fundamental basic research.
Study reference: Qiang Cheng et al. In situ production and secretion of proteins endow therapeutic benefit against psoriasiform dermatitis and melanoma. Proceedings of the National Academy of Sciences. DOI: 10.1073/pnas.2313009120
