Neural networks enlist physics-based computations for faster, clearer image restoration
Fluorescence microscopy allows researchers to study specific structures in complex biological samples. However, the image created using fluorescent probes suffers from blurring and background noise. The latest work from NIBIB researchers and their collaborators introduces several novel image restoration strategies that create sharp images with significantly reduced processing time and computing power1.
The cornerstone of modern image processing is the use of artificial intelligence, most notably neural networks that use deep learning to remove the blurring and background noise in an image. The basic strategy is to teach the deep learning network to predict what a blurry, noisy image would look like without the blur and noise. The network must be trained to do this with large datasets of pairs of sharp and fuzzy versions of the same image. A significant barrier to using neural networks is the time and expense needed to create the large training data sets.
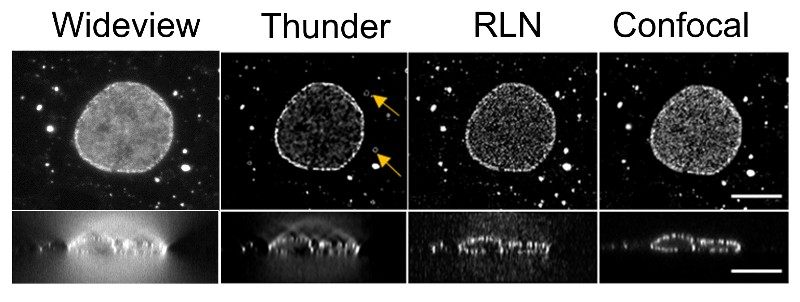
Before the use of neural networks, images were cleaned up—known as deconvolution—using equations. Richardson-Lucy Deconvolution (RLD) employs an equation that uses knowledge of the blurring introduced by the microscope to clear up the image. The image is processed through the equation repeatedly to further improve it. Each pass through the equation is known as an iteration and many iterations are needed to create a clear image. The resources and time that it takes to run an image through many iterations is a main drawback of the RLD approach.
RLD is considered to be physics-driven because it describes the physical properties that cause the blurring and noise in an image. Neural networks are said to be data-driven because they must look at a lot of images (data) to learn what constitutes a fuzzy or clear image. The NIBIB team sought to leverage the advantages—and mitigate the disadvantages—of each method by combining them. The result is a neural network that also uses RLD—a Richardson-Lucy Network (RLN).
By design, a neural network detects characteristics in the matched pairs that will help it learn how to make a fuzzy image clear. Interestingly, the scientists who designed these networks generally do not know the specific characteristics the network is using to accomplish this feat. What is known is that the features detected by the network are based at least in part on physical properties of the microscope and so can be represented by equations.
The team developed a training regimen that incorporates RLD-like equations into the neural network that add information about the physical properties distorting the image. The most helpful equations are recycled through the network--accelerating the learning process. Thus, the iterative equations of RLD were built into the neural network to create RLN.
“We think of this approach as “guiding” the neural network’s learning process,” explains Yicong Wu, Ph.D., a senior author of the study. “Put simply, the guidance helps the network learn more rapidly.”

Tests on images of worm embryos showed that RLN improved both training and processing time compared to other deep learning programs now in use. The number of parameters need to train the network using RLN were dramatically reduced from as many as several million to less than 20,000. The processing time to obtain clear images of the embryos was also greatly reduced with RLN taking only a few seconds to resolve an image compared with 20 seconds to several minutes using other popular neural networks.
Although RLN accelerates the training process, the datasets of fuzzy and clear images needed to train the network are difficult to obtain or create from scratch. To address the problem, the researchers ran RLD equations in reverse to rapidly create synthetic data sets for training. Computer-generated images were created with a mixture of dots, circles, and spheres—termed mixed synthetic data. Based on measurements from out-of-focus cells, blur was added to the synthetic images. Background noise was further added to create blurry, noisy images of the computer-generated synthetic shapes. The pairs of clear and fuzzy synthetic mixed shapes were used to train a neural network to restore actual images of live cells. The experiment demonstrated that RLN trained on synthetic data outperformed RLD in creating clear images of the out-of-focus cells. Impressively, RLN cleared up many fine structures in the images that RLD failed to detect.
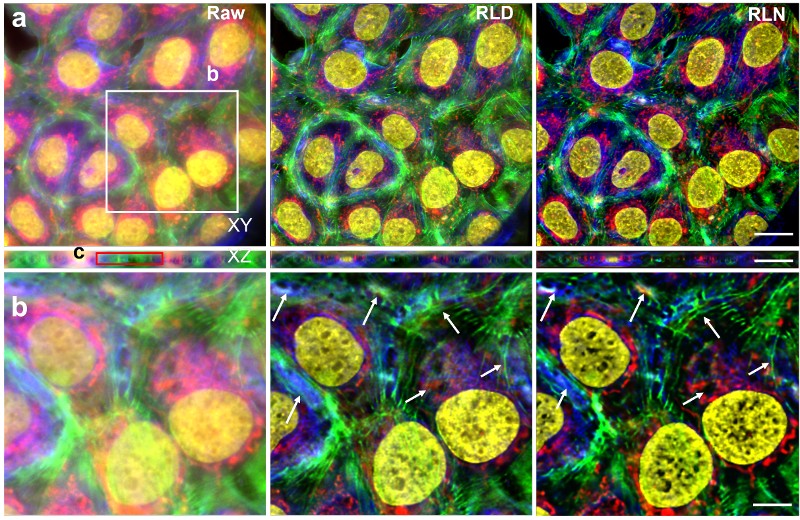
“The success we are having with using synthetic data to train neural networks is very exciting,” explained Hari Shroff, Ph.D., one of the lead authors of the study. “Creating or obtaining data sets for training has been an enormous bottleneck in image processing. This combination of the findings in this work—that synthetic data really works, especially when used with RLN—has the potential to usher in a new era in image processing that we are vigorously pursuing.”
The group is extremely excited about another aspect of the work. They found that synthetic data sets created to restore images of a specific subject such as live cells were also able to restore fuzzy images of completely different images such as human brains. They describe this as the “generalizability” of synthetic training. The team is now moving full speed ahead to see how far such generalizability can be taken to accelerate the creation of high-quality images for biological research.
The work was supported by the intramural laboratories at NIH, the National Institute of Biomedical Imaging and Bioengineering, and the National Institute of Mental Health with additional funding from the NIH Office of the Director, the National Institute of General Medical Sciences and the National Institute of Neurological Disorders and Stroke. Additional support was provided by the National Key Research and Development Program of China, the National Natural Science Foundation of China, the Talent Program of Zhejiang Province, the Marine Biological Laboratories at Woods Hole, and the HHMI. This work used the computational resources of the NIH HPC Biowulf cluster (https://hpc.nih.gov).
-Written by Thomas Johnson, Ph.D.
